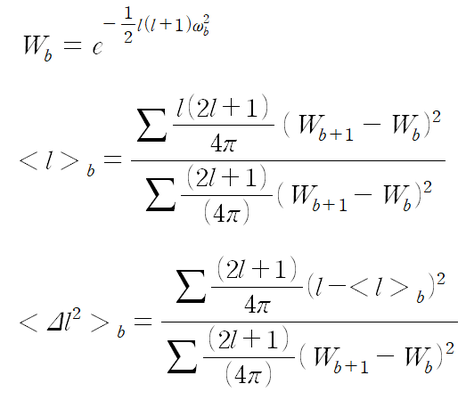Jan 7, 2019 - An attempt at Gaussian filtering (incorrect)
This is my attempt at Gaussian filtering the CIB total map by lbands.
The procedure goes as such:
1. Using healpy.anafast, transform the map into an array of cls.
2. In my instance, the nside was 2048, so lmax was 6144. I removed the last 144 cls and then split the cls into 'nbands' arrays (nbands = 20 here).
3. Generate an 'nbands' by 'nbands' matrix, with the diagonals being the split cls in order, and all other elements '6000/nbands' zeros.
For example, M[0]M[0] = cls[0], M[1]M[1] = cls[1], ... ,M[19]M[19] = cls[19], all other elements: [0, 0, ..., 0] => '6000/nbands' zeros.
4. Flatten each row of the matrix, then create 'nbands' separate maps using healpy.synfast(M[i], 2048).
5. Smooth each of the maps using healpy.smoothing(M[i], fwhm = np.radians(1.0)).
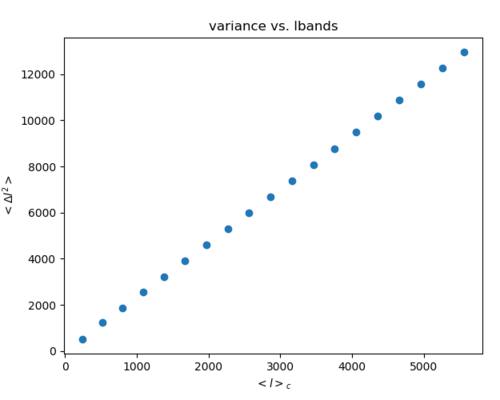
6. Calculate <l>_c and <Delta_l^2> and plot them.
-3 was done so that I could calculate the statistics of each band. I am not completely sure if this is the way to do it.
-There is a catch with using healpy.synfast() because it will produce a different map every time it is run. For example, consider the following: map => anafast => cls => synfast => map1 => cls1 'map' does not equal 'map1' while 'cls' is nearly the same as 'cls1.' So I think there will be some error due to this, but since <Delta_l^2> is the variance of the band and synonymous to the power spectrum, it should not have too large an effect.
This is the result with 20 linearly split bands with a Gaussian beam of fwhm = 1 deg to smooth each map.
Below are the equations used to define parameters with omega being the width (standard deviation) of an l-band in the map while 'W' is the width in l-space: